In this blog post Sri Kashyap and I describe how to deal with the registration of high-resolution datasets across days, across different resolutions, and across different sequences.
I am particularly fond of the following two tools: Firstly, ITK-SNAP for visually-guided manual alignment and secondly, using ANTs programs: antsRegistration and antsApplyTransforms.
Initial manual alignment with ITK-SNAP
ITK-SNAP is a program that can be downloaded here: http://www.itksnap.org/pmwiki/pmwiki.php
In ITK-SNAP, the base image (EPI) and the overlay image (MP2RAGE) are opened separately.
- The transparency can be set between the base and overlay images. Pressing “w” switches the images back and forth.
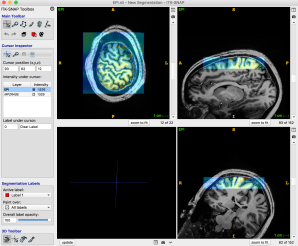
-
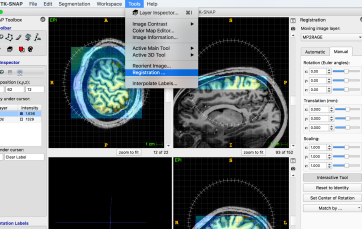
- Select: Tools -> Registration -> Manual
- If there are issue with header information, adjust manually using Interactive Tool
- Then you go to automatic tab and run Registration. I liked the following parameters:
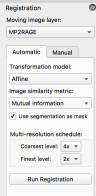 However, note that this does not always work. one can try rigid and other similarity metrics. For instance, when the FOV is too small, the automatic registration can sometimes put the slab out of place. In this case, the manual adjustments can suffice. However, this approach has worked for single slice fMRI studies (see below).
However, note that this does not always work. one can try rigid and other similarity metrics. For instance, when the FOV is too small, the automatic registration can sometimes put the slab out of place. In this case, the manual adjustments can suffice. However, this approach has worked for single slice fMRI studies (see below). - save the transformation file in ITK format with save symbol

- Now you can apply it with command:
-
antsApplyTransforms --interpolation BSpline[5] -d 3 -i MP2RAGE.nii -r EPI.nii -t initial_matrix.txt -o registered_applied.nii
High-quality registration with ANTS
- ANTS documentation: https://github.com/stnava/ANTsDoc/raw/master/ants2.pdf
antsRegistration \ --verbose 1 \ --dimensionality 3 \ --float 1 \ --output [registered_,registered_Warped.nii.gz,registered_InverseWarped.nii.gz] \ --interpolation Linear \ --use-histogram-matching 0 \ --winsorize-image-intensities [0.005,0.995] \ --initial-moving-transform initial_matrix.txt \ --transform Rigid[0.05] \ --metric CC[static_image.nii,moving_image.nii,0.7,32,Regular,0.1] \ --convergence [1000x500,1e-6,10] \ --shrink-factors 2x1 \ --smoothing-sigmas 1x0vox \ --transform Affine[0.1] \ --metric MI[static_image.nii,moving_image.nii,0.7,32,Regular,0.1] \ --convergence [1000x500,1e-6,10] \ --shrink-factors 2x1 \ --smoothing-sigmas 1x0vox \ --transform SyN[0.1,2,0] \ --metric CC[static_image.nii,moving_image.nii,1,2] \ --convergence [500x100,1e-6,10] \ --shrink-factors 2x1 \ --smoothing-sigmas 1x0vox \ -x mask.nii
Note the “initial_matrix.txt” here is from ITK-SNAP and is used to get a good initialisation for ANTs.
The 3/4 after -d means the dimension, use either 3 or 4 dimensional registration -n means the number of cores. -x is a flag defining a mask for the registration optimisation.
You can always apply the same spatial transformation with “antsApplyTransforms”:
antsApplyTransforms --interpolation BSpline[5] -d 3 -i moving_image.nii -r reference_image.nii -t registered_1Warp.nii.gz -t registered_0GenericAffine.mat -o registered_applied.nii
The order of applying transforms is important. More information about ANTS details here: https://github.com/ANTsX/ANTs/wiki/Anatomy-of-an-antsRegistration-call
Note here that the T1w map from the MP2RAGE was registered to the EPI images. This is in contrast to standard analysis in low-resolution fMRI studies, where the anatomy was registered to the functional data in order to keep the functional data in its original space and, hence, avoid any additional smoothing.
Video Tutorial
The following video tutorial goes through all the steps above on example data, that can be downloaded here.
Examples of registration
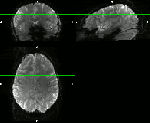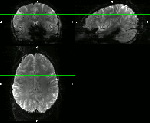
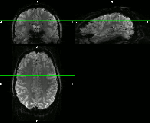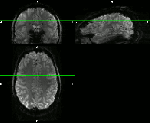
These are examples from the area of DLPFC (dorso lateral prefrontal cortex), an area investigated from Emily Finn in this study. Here, different fMRI protocols were used across multiple MRI sessions for the same task. The goal of the registration was to see whether the same ROI was detected for different fMRI protocols with different coverage and resolution:
- ‘Conventional’ 2mm whole brain fMRI with TR=1s across different days. (click on the figures to enhance)
- Test-retest experiments with an axial-like sub-millimeter slab.
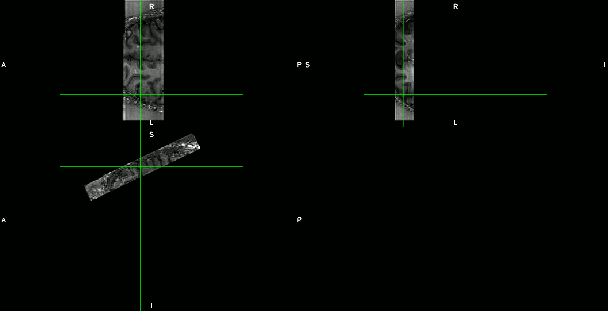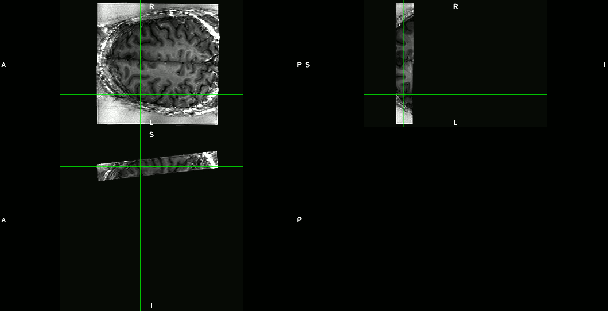
- Axial-like and sagittal high-resolution slabs. (click on the figures to enhance)
ANTS registration of single slice data à la Sri
The registration with ANTS is also helpful for very unconventional data. e.g. single slice data, which is not really supported by many alternative software suites.
To achieve 0.1mm (anisotropic) resolutions, in this paper, the coverage was limited to a single slice. Thus, here the high resolution T1 map from the MP2RAGE (acquired with the same orientation as the single-slice data) was registered to the single-slice fMRI mean image using the workflow similar to above and is described in the supplementary material of the paper.
The two-step registration process using the initial matrix from ITK-SNAP together with antsRegistration command for registration is the backbone to achieving highly accurate registration of very high resolution, unconventional data types.
The scripts and example data of single slice fMRI will be made available as soon as I get them from Sri.



Is there a way to transform the layer fMRI into MNI152 space? Or the transformation from MNI152 to the layer fMRI?
LikeLike
Sure, just use the MNI-atlas as your “static_image.nii” .
Make sure you use the T1-weighted contrast. So it matches the EPI data.
LikeLike
Hello sir, thank you for such greatfull informations. I want to ask if it’s possible to get the source code for regestration,pls. Or are there any suggestion?
LikeLiked by 1 person
Sure, all the scripts that I used here are available on: https://github.com/layerfMRI/repository/tree/master/Alignement_scripts
Those scripts call ANTS, the corresponding source code is available here: https://github.com/ANTsX/ANTs
LikeLike
Hello, thanks for your excellent sharing. I want to ask if it’s possible to get the source code for regestration of single slice data. Thank you!
LikeLike