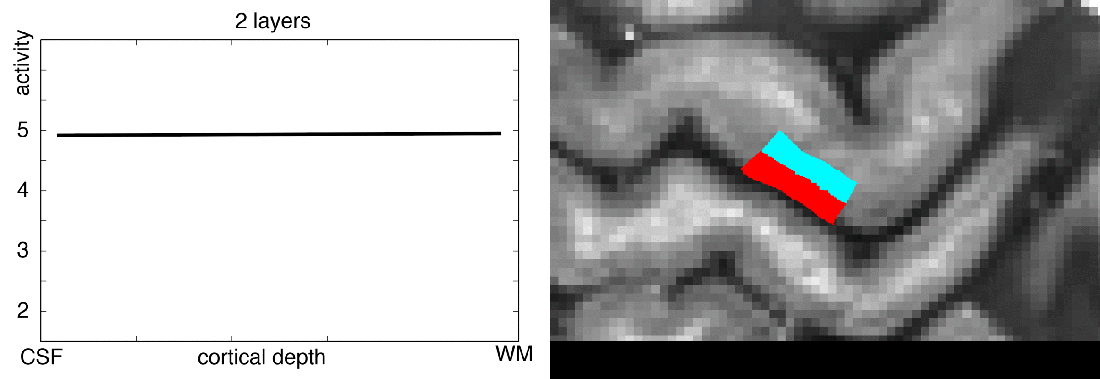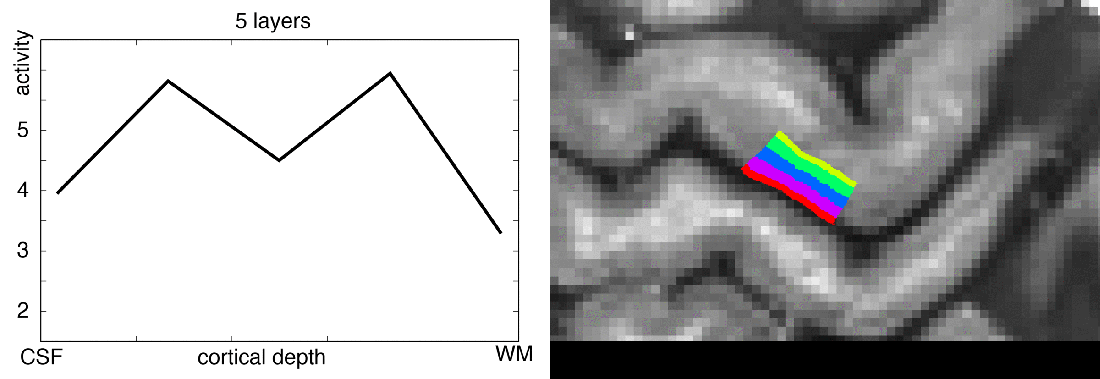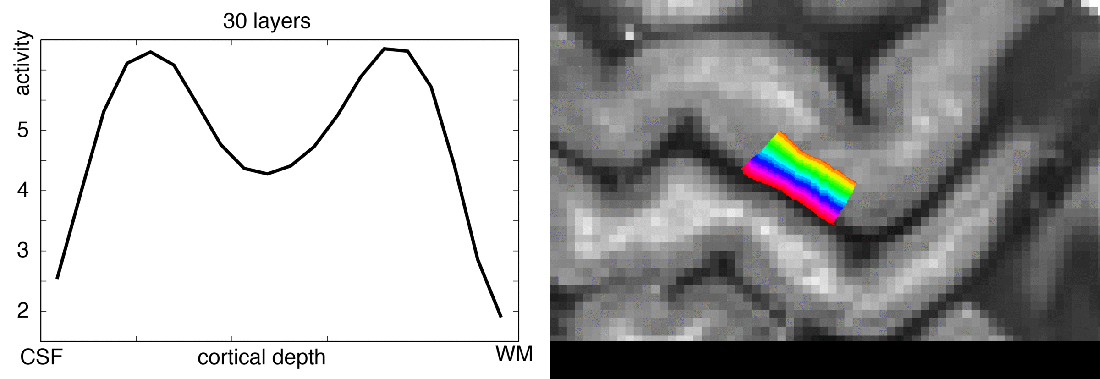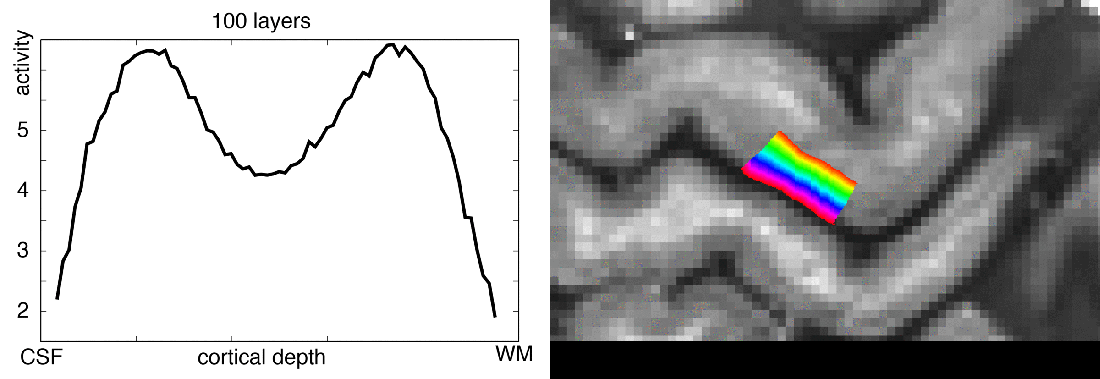
Meeting minutes of Study group on brain functions business meeting:
15:30-16:30 in W07, June 21st, 2018, ISMRM, Paris:
Chunlei Liu presents the talk: “Ultra-high resolution fMRI – from hardware to pulse sequences to the human brain – and vice-versa. A 7T BRAIN Initiative Project” per pro David Feinberg.
In the subsequent Q/A period it is discussed that the current gradient design will be “asymmetric” and that the target parallel-imaging acceleration factor will be 20-30.
Past Chair Essa Yacoub presents the study group statistics:
There are many more trainees than full members.
Natalia Petridou presents the Trainee abstract awards:
- #1 Adam Yamamoto for the abstract entitled: Intra-operative acquisition of sensorimotor fMRI during glioma resection: evaluation of feasibility and clinical applicability.
- #2 Domenic Cerri for the abstract entitles: Mechanisms underlying negative fMRI response in the striatum. Since Dominic could not make it, he sent one of his collogues to receive the price for him.
- #3 Yi-Tien Li for the abstract entitled: Inter-Regional BOLD Latency after Vascular Reactivity Calibration is Correlated to Reaction Time.
Renzo Huber presents the results of the survey among the study group members “Biggest challenges of high-resolution fMRI”. The slides can be downloaded here: https://doi.org/10.7490/f1000research.1115658.1
The attendees of the business meeting commented on the biggest challenges of high-resolution fMRI and the issues that could be discussed in a future virtual study group meeting.
· Unknown (to me) senior study group member: At very high resolutions, the elastography of the brain might become a limit of submillimeter fMRI. This comment might be related to the Tweet from NIH Director Francis Collins that went viral on social media earlier that day: https://twitter.com/NIHDirector/status/1009793641550893056 (with amplified motion).
· Ravi Menon: Motion is a big limitation of sub-millimeter fMRI.
· Cheryl Olman: Every person/group uses a different analysis pipeline. It would be helpful to have a synthetic standard dataset that is open to everyone. Every person could play around with it and test the quality of various evaluation pipelines.
· Hanzhang Lu: The virtual study group meeting would lose the interest of many study group members if the focus is only of 7T imaging. It would be helpful to also discuss the spatial specificity limitations at 3T too.
· Shella Keilholz: Since there are so many trainee members, they should mention what they need.
· Olivia Stanley (“encouraged” from Ravi Menon):
- Trainees want to know what segmentation tools are out there and how to use them.
- Trainees want more information on registration methods.
- Trainees would benefit from a tutorial on how to share code and how to use shared code.
· Ravi Menon: So many people are sharing their code on various platforms. It would be helpful to have an online repository of repositories.
· Essa Yacoub: It will be hard to standardize analysis pipelines because the data and the acquisition hardware is not standardized.
· Trainee member (not known to me): The analysis of sub-millimeter fMRI takes a lot of computational recourses. Trainees can be limited by this.
· Nikolaus Weiskopf: The image reconstruction can be a black box. It is not straight-forward to share pipelines of the Image reconstruction.
· Nikolaus Weiskopf: Data-sharing becomes more difficult in the EU because of increasingly stricter privacy protection regulations.
· Natalia Petridou: Data sharing is increasingly encouraged from the funding agencies and the journals.
· James Pekar: The field of sub-millimeter fMRI stands on the shoulder of decades of preceding work from volunteers and people with passion. This study group can help todays volunteers and people with passion to increase their visibility.